Derm-T2IM: Harnessing Synthetic Skin Lesion Data via Stable Diffusion Models for Enhanced Skin Disease Classification using ViT and CNN

Introduction
Skin cancer remains a significant global health concern, with its incidence rising due to factors such as increased ultraviolet (UV) exposure, aging populations, and lifestyle changes. Early detection is crucial for effective treatment, and artificial intelligence (AI) has become an invaluable tool in assisting dermatologists with diagnosing skin diseases, including melanoma and other malignancies. However, developing robust AI models necessitates large, diverse, and high-quality datasets of skin lesion images—a requirement often hindered by challenges like privacy regulations, data variability, and the need for expert annotations.
To address these limitations, researchers are turning to synthetic data generation. By creating artificial skin lesion images that closely resemble real ones, it's possible to augment existing datasets, thereby enhancing the training of AI models. The study titled "Derm-T2IM: Harnessing Synthetic Skin Lesion Data via Stable Diffusion Models for Enhanced Skin Disease Classification using ViT and CNN" introduces an innovative approach to this problem. The researchers employ stable diffusion models—a type of generative AI—to produce high-quality synthetic skin lesion images, aiming to improve the performance of skin disease classification systems.
In this blog post, we'll delve into the Derm-T2IM study, exploring how this method works and its potential implications for medical imaging and AI-driven diagnostics.
Overview of the Proposed Methodology
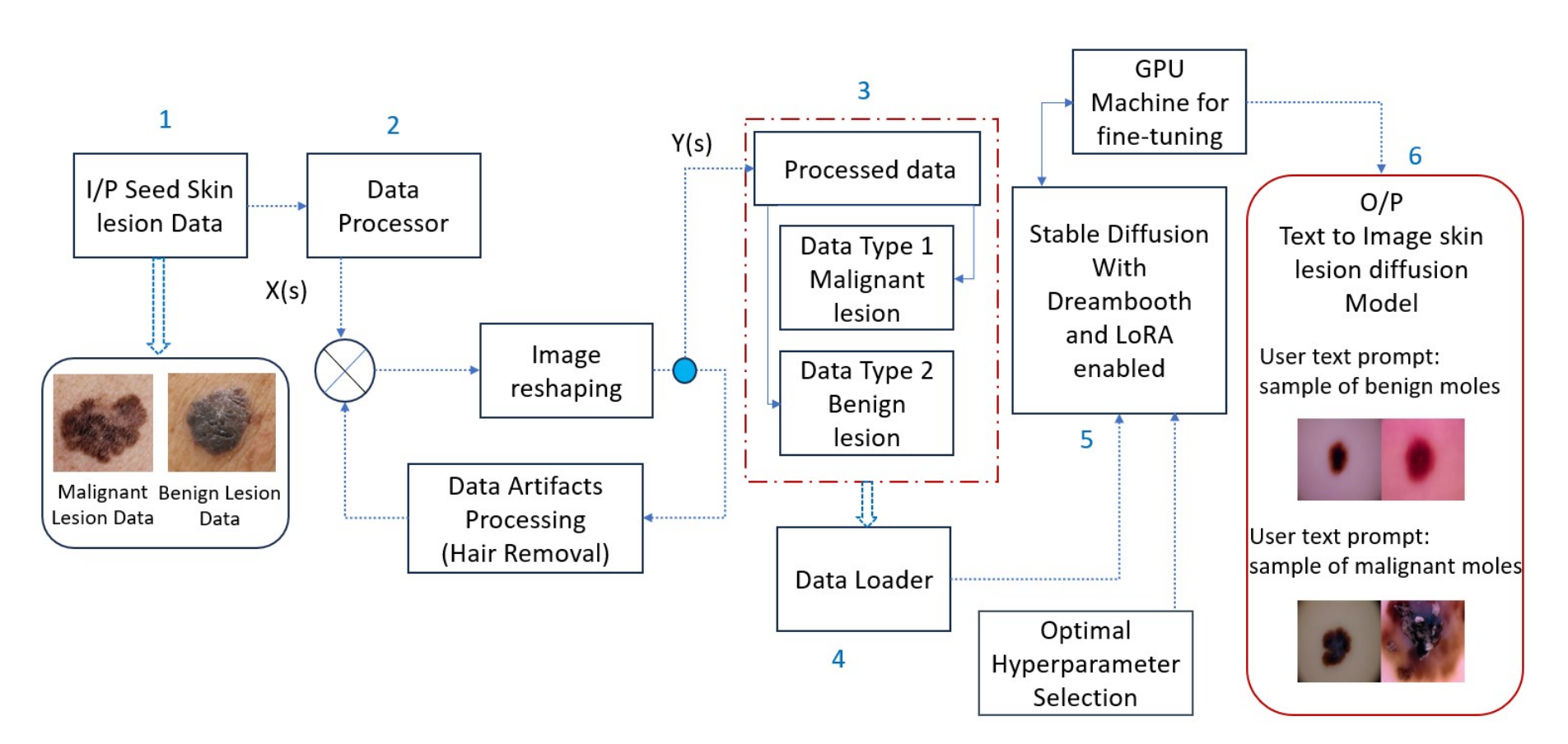
Before exploring the specific components of Derm-T2IM, it’s useful to understand the overall workflow of the proposed methodology. Many steps while building DERM-T2IM are shown in the diagramm but there are overall three mains parts. The first part is data processing and data loading for training the model. For this paper, the author built Derm-T2IM to generate malignant and benign lesions images. Data preprocessing incorporates resizing the image size to 512 × 512 as input tensor shape and further removing data clutters such that unwanted skin hairs covering the lesion region of interest thus properly focusing on lesion area.The processed data is divided in two different data loading concepts which includes malignant skin lesion data and benign skin lesion data. The second part is the training process which uses Stable diffusion with Dreambooth and LoRA enabled.Finally, once the model is trained, various image sampling methods are used to render new synthetic dermatoscopic data.
Components of Derm-T2IM
Derm-T2IM, short for Dermatoscopic Text-to-Image Model, is a customized framework built on stable diffusion technology, designed to generate synthetic skin lesion images from text prompts. Let’s break down its key components:
- Pre-trained Stable Diffusion Model
At its core, Derm-T2IM starts with a pre-trained stable diffusion model. These models are generative AI systems that create realistic images by reversing a process where noise is gradually added to data. In this case, the model takes text descriptions (e.g., “malignant skin lesion”) and generates corresponding images, forming the foundation of Derm-T2IM. - DreamBooth for Few-Shot Learning
To tailor the model to skin lesions, the researchers use Dreambooth, which is being developed by Google research and Boston University researchers offers the unique advantage of utilizing a small set of seed training data depicting a particular data class/subject, by adapting a pre-trained textto-image Imagen model to associate a distinct identifier with that specific subject. After integrating the subject into the model’s output space, this identifier becomes a tool for generating new images of the subject within various contextual settings. The approach leverages the inherent semantic knowledge within the model, along with a novel self-generated class-specific prior preservation loss, thus allowing to create representations of the subject in a wide range of scenes, poses, perspectives, and lighting conditions that were not present in the original reference images. - Low-Rank Adaptation (LoRA)
Fine-tuning large models can be computationally intensive, so Derm-T2IM employs Low-Rank Adaptation (LoRA). This method optimizes the model efficiently by adjusting only a small subset of parameters, reducing training time and resource demands while maintaining performance. - Image Inference Samplers
After training the model using few-shot learning and optimized parameters, synthetic dermatoscopic images are generated using three text-to-image sampling methods: Euler, Euler a, and PLMS. Euler is a deterministic method similar to solving differential equations without adding noise. Euler a introduces controlled random noise during sampling, making it an ancestral method where each step depends on previous noise. PLMS (Pseudo Numerical Methods for Diffusion Models on Manifolds) approximates diffusion processes on manifolds to generate realistic data by simulating the dynamics of complex data distributions through numerical steps.
Experiments and Results
The researchers put Derm-T2IM through a rigorous development and testing process. Here’s how it unfolded:
- Fine-Tuning Process
Derm-T2IM was fine-tuned using 2800 images (1400 benign, 1400 malignant) from the ISIC Archive. The training ran for 120 epochs with a batch size of 2, using the AdamW optimizer and a learning rate of 2e-6. Mixed precision (FP16) kept memory usage low, completing the process in 10.4 hours on an RTX 3090Ti GPU. The loss steadily decreased, hitting 0.1394, signaling effective learning. - Image Generation
Post-tuning, Derm-T2IM generated synthetic images using three sampling methods: Euler, Euler a, and PLMS. These methods determine how the model constructs images from noise, with sampling steps between 20-26 yielding high-quality 512x512 PNG outputs. The classifier-free guidance (CFG) scale was set to 7, balancing fidelity to text prompts. - Smart Transformations
The model’s flexibility shone through in its ability to produce “smart transformations.” By varying text prompts, Derm-T2IM created images with specific traits—like different mole sizes, multiple moles, or lesions on diverse skin tones—demonstrating its adaptability for tailored data needs.
The result? A dataset of 6000 synthetic images (3000 benign, 3000 malignant), open-sourced for the research community via GitHub, alongside the Derm-T2IM model on Hugging Face.
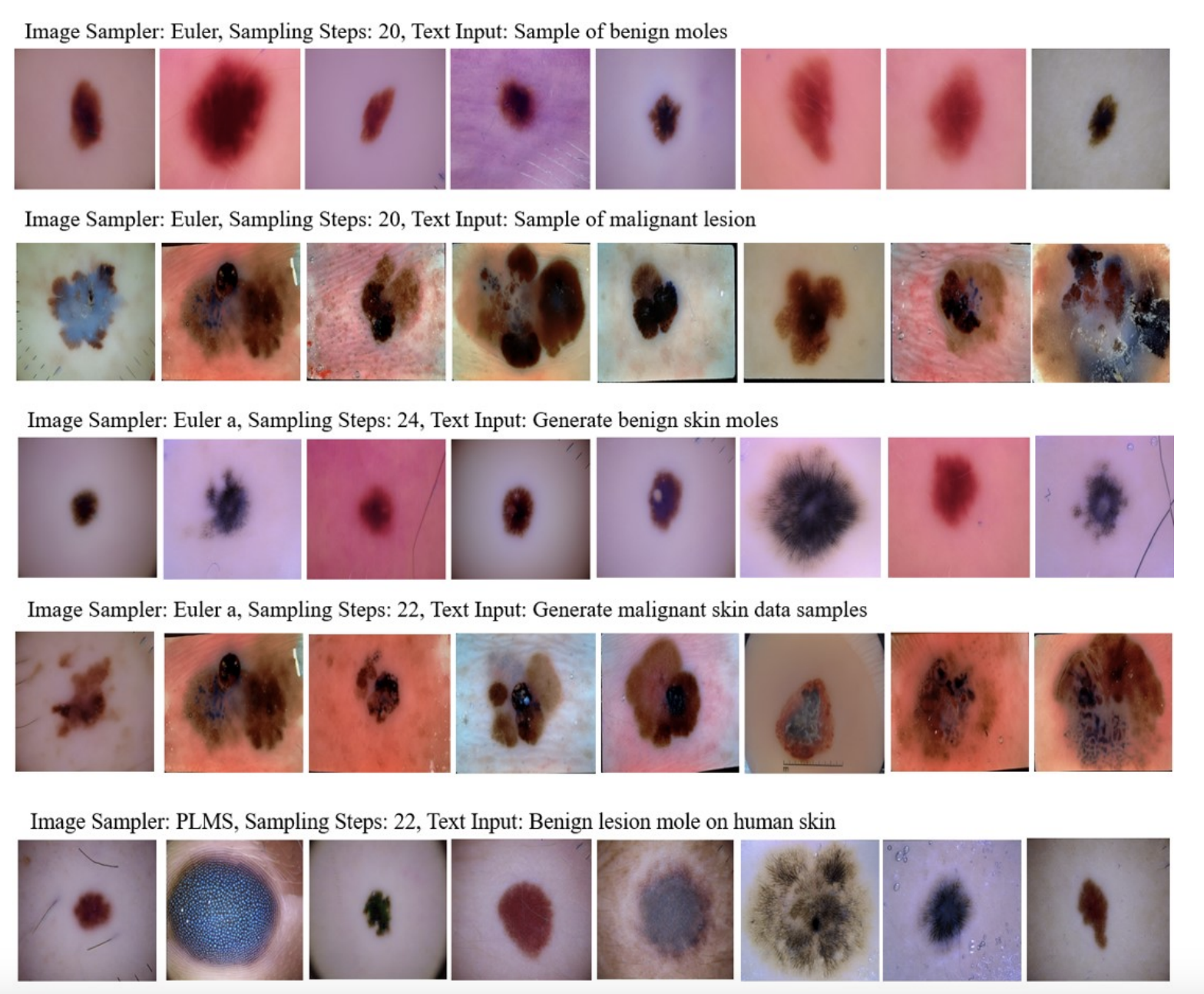
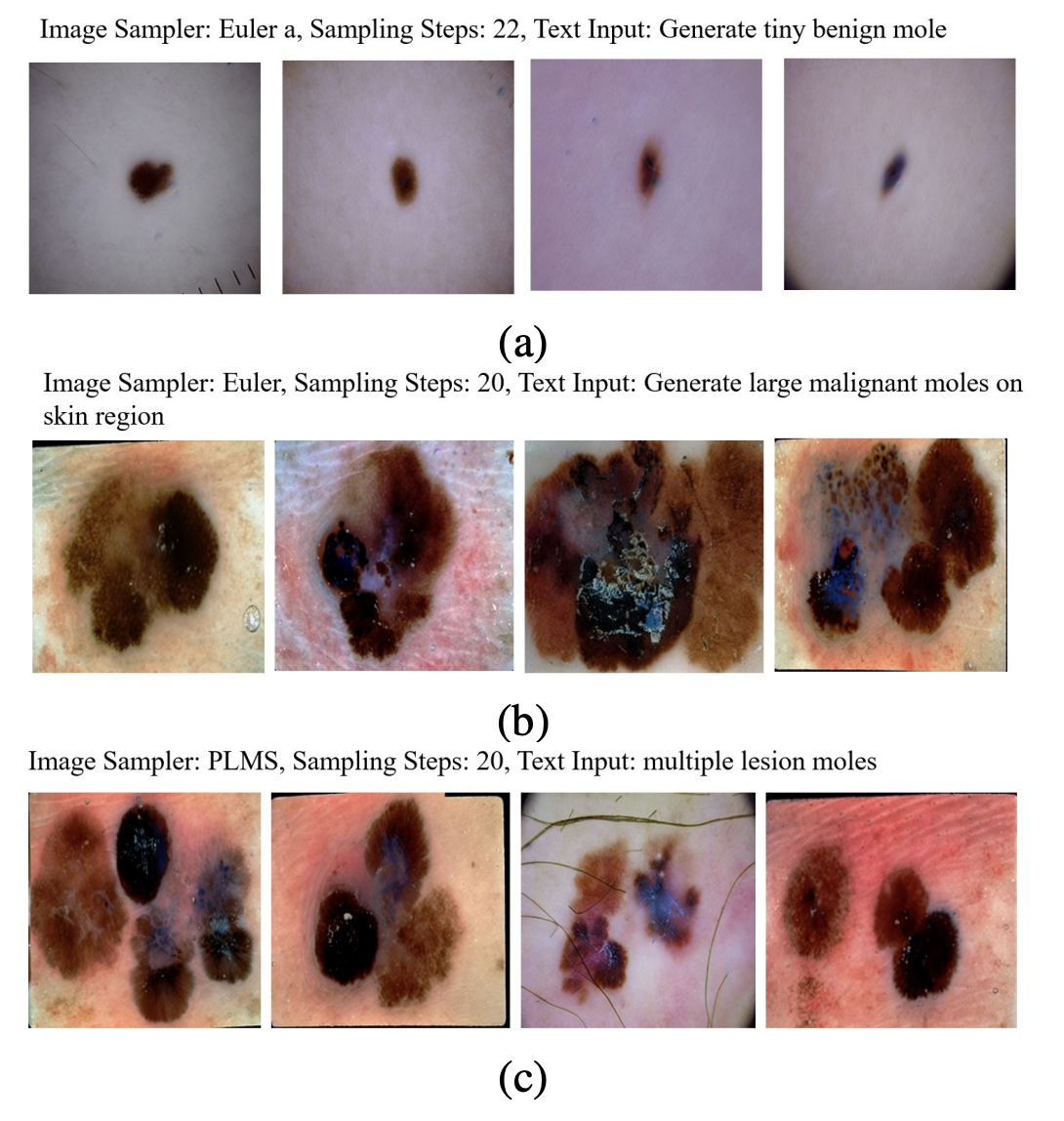
Synthetic Data Validation
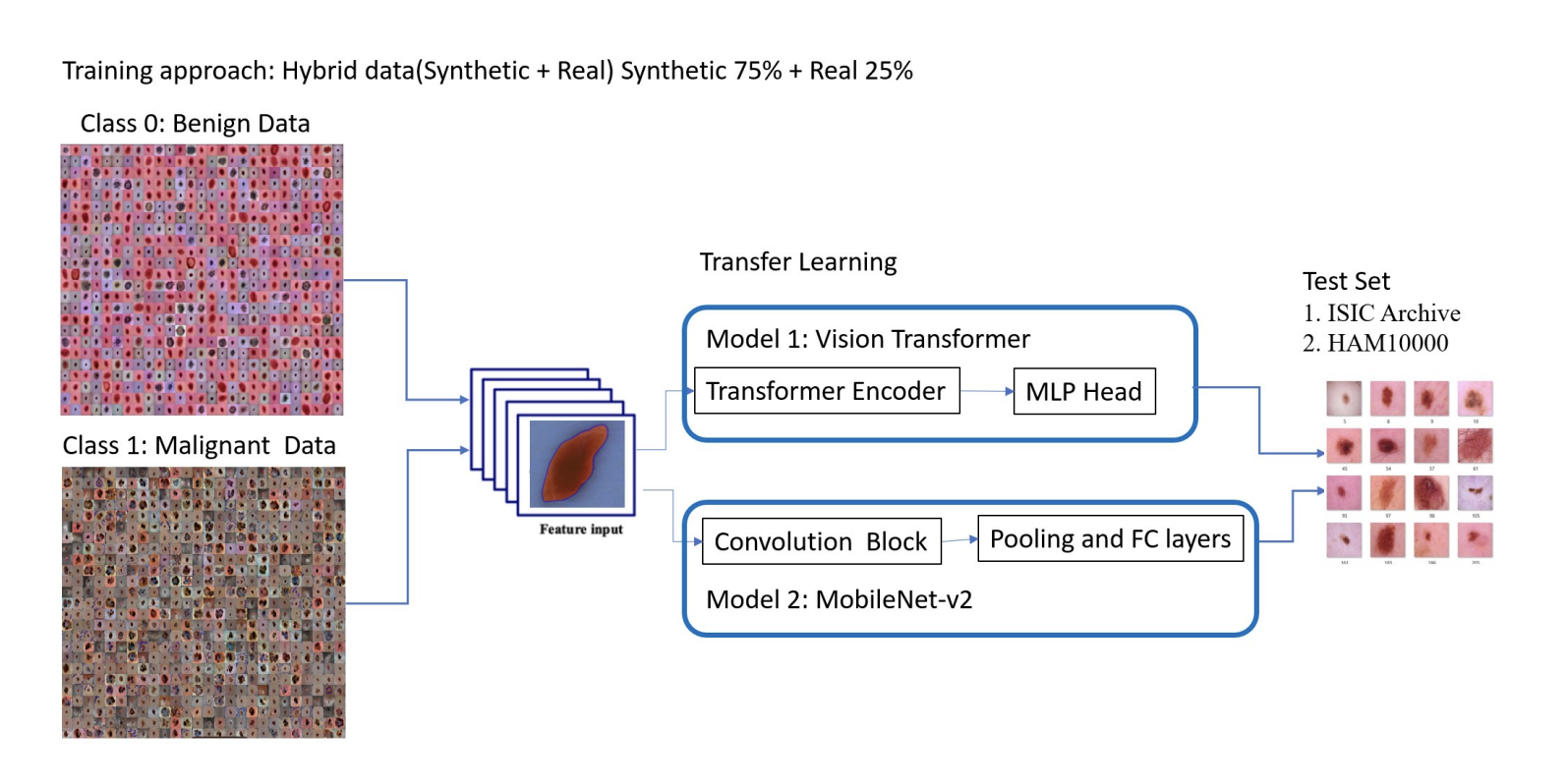
To prove the synthetic data’s worth, the researchers tested its impact on two classification models: a Vision Transformer (ViT) and a MobileNet V2 CNN. Here’s how they validated it:
- Hybrid Training Approach
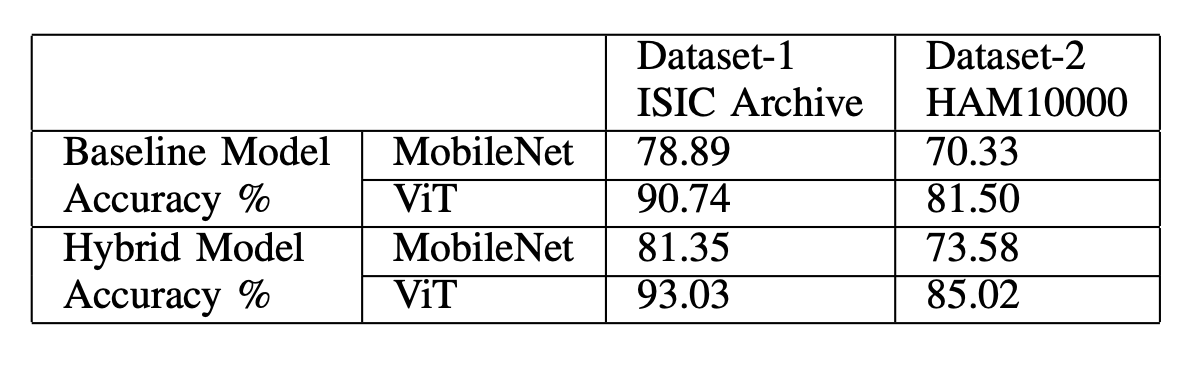
They trained the classifiers using a mix of 75% synthetic and 25% real data, comparing this hybrid approach to baselines trained solely on real data. The hybrid models were then tested on unseen real-world datasets: ISIC Archive and HAM10000.
- Performance Gains
The results were striking:- ISIC Archive: ViT accuracy rose from 90.74% (real data only) to 93.03% (hybrid), while MobileNet V2 improved from 78.89% to 81.35%.
- HAM10000: ViT jumped from 81.50% to 85.02%, and MobileNet V2 from 70.33% to 73.58%.
These boosts highlight how synthetic data enhances model generalization.
- Qualitative Checks
Beyond classification, they validated the data’s realism:- Zero-Shot Segmentation: The Segment Anything model segmented moles in synthetic images accurately without additional training, confirming their structural fidelity.
- Detection and Classification: A pre-trained YOLO-V8 model, tuned on real ISIC data, detected and classified moles in 200 synthetic images with 72% accuracy, further validating their quality.
These tests affirm that Derm-T2IM’s synthetic data is not just realistic but practically useful for downstream tasks.
Conclusion
The Derm-T2IM study demonstrates a groundbreaking use of stable diffusion models to tackle the data scarcity problem in medical imaging. By combining few-shot learning with efficient fine-tuning, the researchers crafted a tool that generates diverse, high-quality synthetic skin lesion images. When paired with real data, these images significantly boost the performance of skin disease classifiers, offering a scalable solution to challenges like privacy concerns and limited dataset diversity.
The open-sourcing of Derm-T2IM and its 6000-image dataset is a gift to the research and medical communities, paving the way for further innovation. Looking ahead, the model could be expanded to cover more skin conditions—like acne or burns—or refined for even greater user control over image generation. Ultimately, Derm-T2IM underscores the transformative potential of synthetic data in advancing AI-driven diagnostics, promising better tools for early detection and improved patient outcomes.
References
- Muhammad Ali Farooq, Wang Yao, Michael Schukat, Mark A Little, Peter Corcoran Derm-T2IM: Harnessing Synthetic Skin Lesion Data via Stable Diffusion Models for Enhanced Skin Disease Classification using ViT and CNN https://arxiv.org/pdf/2401.05159
- Robin Rombach, Andreas Blattmann, Dominik Lorenz, Patrick Esser, Björn Ommer High-Resolution Image Synthesis with Latent Diffusion Models https://arxiv.org/pdf/2112.10752
- Nataniel Ruiz, Yuanzhen Li, Varun Jampani, Yael Pritch, Michael Rubinstein, Kfir Aberman DreamBooth: Fine Tuning Text-to-Image Diffusion Models for Subject-Driven Generation https://arxiv.org/pdf/2208.12242
