MAXVIT-UNET: MULTI-AXIS ATTENTION FOR MEDICAL IMAGE SEGMENTATION
Since their emergence, Convolutional Neural Networks (CNNs) have significantly advanced medical image analysis but struggle with capturing global interactions. Transformers excel in processing global features but face scalability and bias issues. To address these issues, hybrid vision transformers (CNN-Transformers) have been developed, combining the strengths of both convolution and self-attention mechanisms. Inspired by the results of Multi-Axis self-attention (Max-SA), the researchers have used its potential for medical image segmentation to develop a novel architecture dubbed MaxViT-UNet, which uses a UNet-style framework.
MaxViT: Multi-Axis Vision Transformer
MaxVit is a new hybrid architecture which effectively blends the multi-axis attention with convolutions, presented by Google research in 2022.
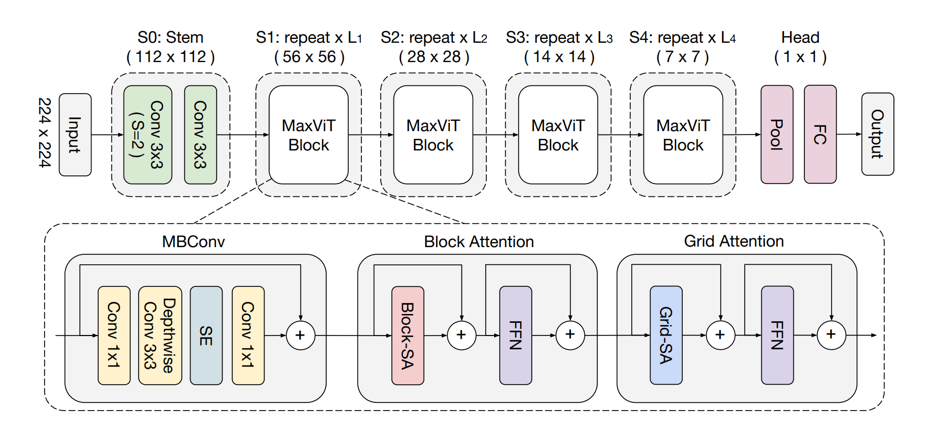
Apparently, the MaxVit architecture is likely a CNN architecture with many feed-forward blocks, decreasing size feature maps, and a fully connected (FC) classification head. The key differentiator is the MaxViT block, which consists of three sub-blocks: Mobile Inverted Bottleneck Convolution (MBConv), Block Attention, and Grid Attention. MBConv is a CNN block used in many architectures such as MobileNet and EfficientNet. Block Attention and Grid Attention serve the role of self-attention.
MBConv
Experiments have shown that MBConv, together with attention, further increases the generalization and trainability of the network. Using MBConv layers prior to attention offers another advantage: depthwise convolutions can be regarded as conditional position encoding (CPE), making the model free of explicit positional encoding layers.
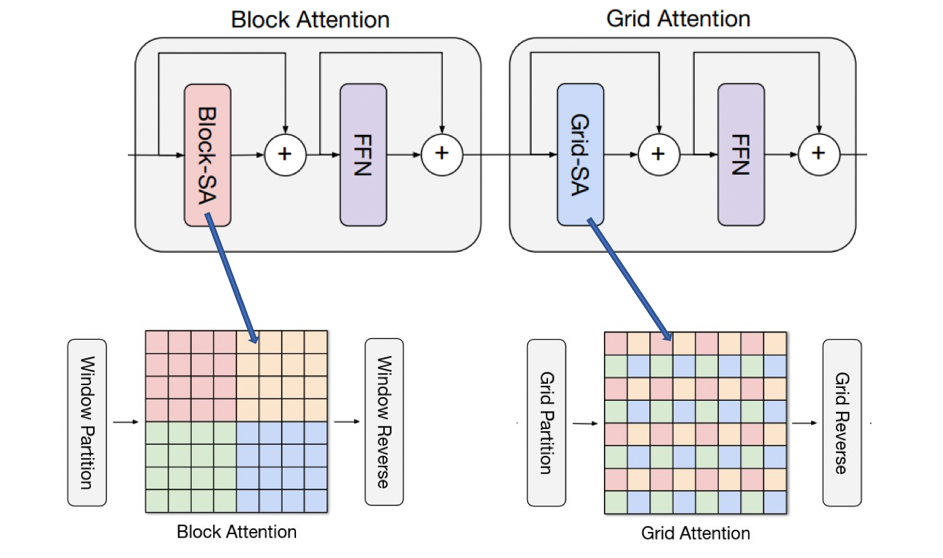
Block Attention
This sub-block conducts local interactions. Let X ∈ R H×W×C be an input feature map. Instead of applying attention on the flattened spatial dimension HWHWHW, we partition the feature into a tensor of shape shape ( H × W/ P × P , P × P, C), representing non-overlapping windows of size P×P. Applying self-attention on the local spatial dimension, i.e., P×P, is equivalent to attending within a small window. This sub-block also adopts typical Transformer designs, including LayerNorm, feedforward networks (FFNs), and skip connections.
Grid Attention
This sub-block aims to gain sparse global attention. Instead of partitioning feature maps using a fixed window size, we grid the tensor into the shape (G×G,H×W/G×G,C), using a fixed G×G uniform grid, resulting in windows of adaptive size H/G×W/G. Employing self-attention on the decomposed grid axis, i.e., G×G, corresponds to dilated, global spatial mixing of tokens.
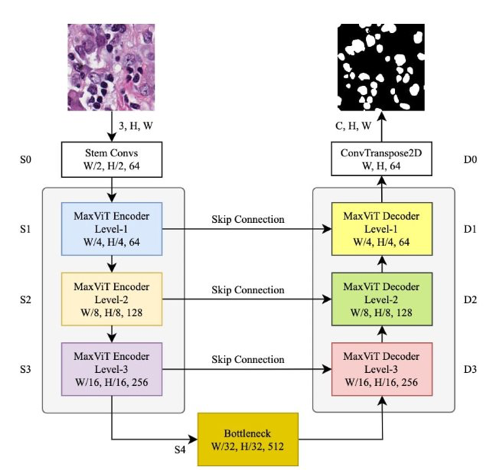
MaxVit-Unet
Inspired by the results of Multi-Axis self-attention, the authors have used its potential for medical image segmentation to develop a novel architecture dubbed MaxViT-UNet, which uses a UNet-style framework. The encoder generates hierarchical features at four scales. The proposed decoder first upscale the bottom-level features, merges them with skip-connection features, and applies the MaxViT-based hybrid feature processing blocks a couple of times to produce an output mask image for the "C" number of classes.
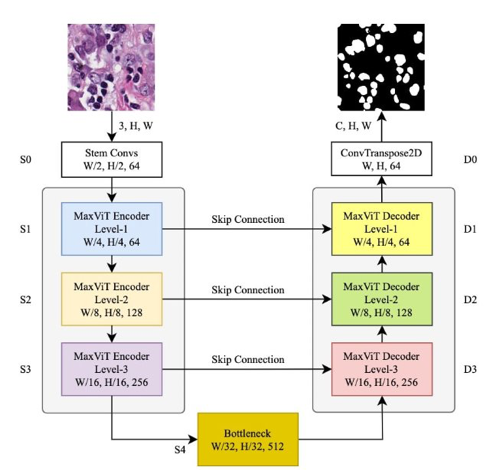
The proposed MaxViT-UNet, though hybrid in nature, consists of only 24.72 million parameters, making it lighter than UNet with 29.06 million parameters and Swin-UNet with 27.29 million parameters. In terms of computation, the proposed MaxViT-UNet takes 7.51 GFlops compared to UNet and Swin-UNet, which take 50.64 and 11.31 GFlops, respectively.
Experiments and Results
Extensive experiments were conducted on a nuclear segmentation task to illustrate the effectiveness of the proposed Hybrid Decoder Network and the MaxViT-UNet segmentation framework. Two datasets, MoNuSeg18 and MoNuSAC20, were used for binary semantic segmentation and multi-class semantic segmentation problems, respectively.
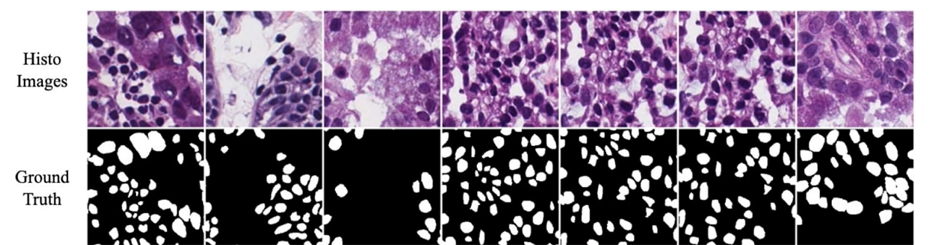
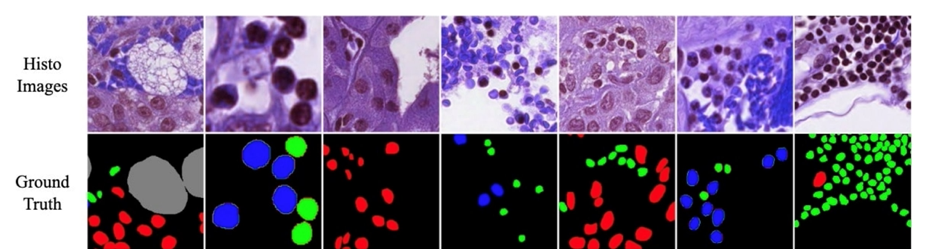
This is the comparison between the proposed MaxViT-UNet and previous methodologies on the MoNuSeg18 dataset and on the MoNuSAC20 dataset
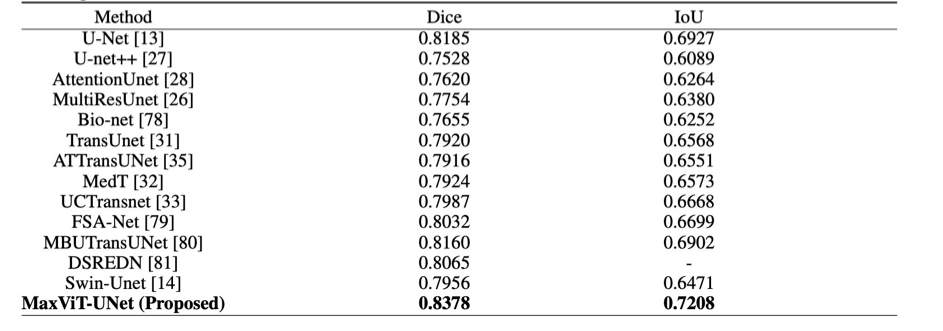
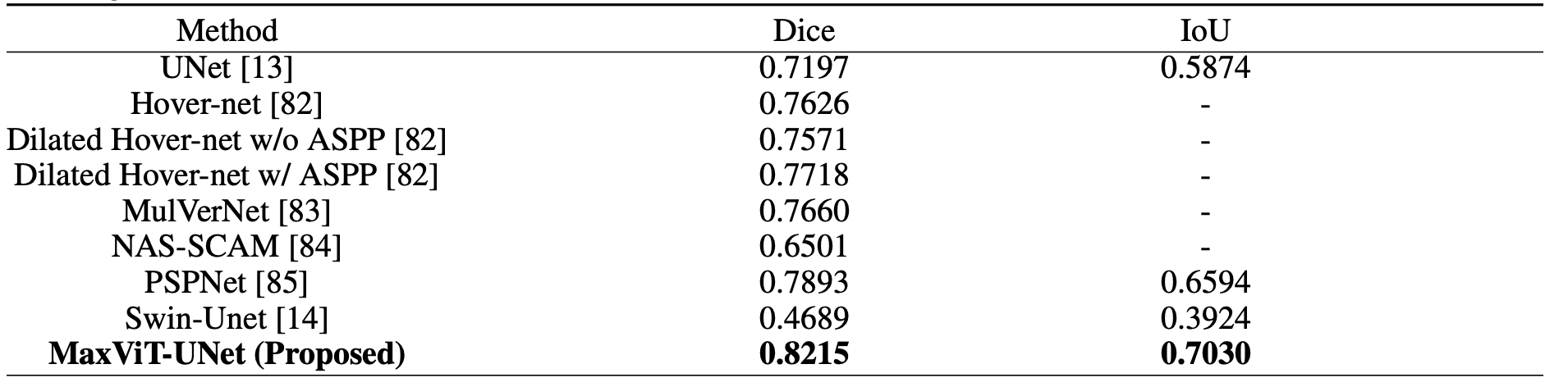
The proposed MaxViT-UNet beats the previous techniques by a large margin on both datasets and proves the significance of the hybrid encoder-decoder architecture.
On MoNuSeg18 dataset, the CNN-based UNet achieved 0.8185 Dice and 0.6927 IoU scores. Whereas, the Transformerbased Swin-Unet achieved 0.7956 Dice and 0.6471 IoU scores. In comparison, our proposed hybrid framework MaxViT-UNet is able to achieve superior scores for both Dice (0.8378) and IoU (0.7208) metrics. It surpassed the CNN-based UNet by 2.36% Dice score and 4.06% IoU score; and Transformer-based Swin-UNet by 5.31% Dice score and 11.40% IoU score on MoNuSeg18 dataset.
For the MoNuSAC20 dataset, the CNN-based UNet achieved 0.7197 mDice (mean Dice) and 0.5874 mIoU (mean IoU) scores. Whereas, the Transformer-based Swin-Unet achieved 0.4689 Dice and 0.3924 IoU scores. In comparison, our proposed hybrid framework MaxViT-UNet is able to achieve superior scores for both Dice (0.8215) and IoU (0.7030) metrics. It surpassed CNN-based UNet by 14.14% Dice and 19.68% IoU scores; and Transformer-based Swin-UNet by a large margin on Dice and IoU metrics as evident from Table 4. The large improvement in both mDice and mIoU scores shows the significance of hybrid encoder-decoder architecture.
Conclusion
MaxViT-UNet represents a promising approach for medical image segmentation, combining the advantages of CNNs and Transformers to achieve superior performance with reduced computational requirements. This hybrid model paves the way for more efficient and effective medical image analysis, ultimately contributing to advancements in medical diagnostics and treatment planning.
References
- Abdul Rehman Khan , Asifullah Khan. MaxViT-UNet : attention multi-axes pour la segmentation des images médicales. https://arxiv.org/pdf/2305.08396
- Zhengzhong Tu, Hossein Talebi, Han Zhang, Feng Yang, Peyman Milanfar, Alan Bovik, and Yinxiao Li. Maxvit: Multi-axis vision transformer. In Computer Vision–ECCV 2022: 17th European Conference, Tel Aviv, Israel, October 23–27, 2022, Proceedings, Part XXIV, pages 459–479. Springer, 2022.
