Revolutionizing Cancer Diagnosis with the CHIEF Foundation Model
Introduction
Cancer diagnosis and prognosis prediction have long relied on histopathology image evaluation, a labor-intensive process that requires expert pathologists to analyze whole slide images (WSIs). With the advancement of artificial intelligence (AI), histopathology has seen tremendous progress, yet existing AI models face significant limitations. Traditional deep learning approaches often require large datasets for training, struggle with tissue diversity, and overfit when trained on limited sources. Addressing these challenges, a new foundation model, CHIEF (Clinical Histopathology Imaging Evaluation Foundation model), has emerged as a groundbreaking AI system designed to improve cancer diagnosis and prognosis prediction.
The CHIEF Framework: A General-Purpose Pathology AI System
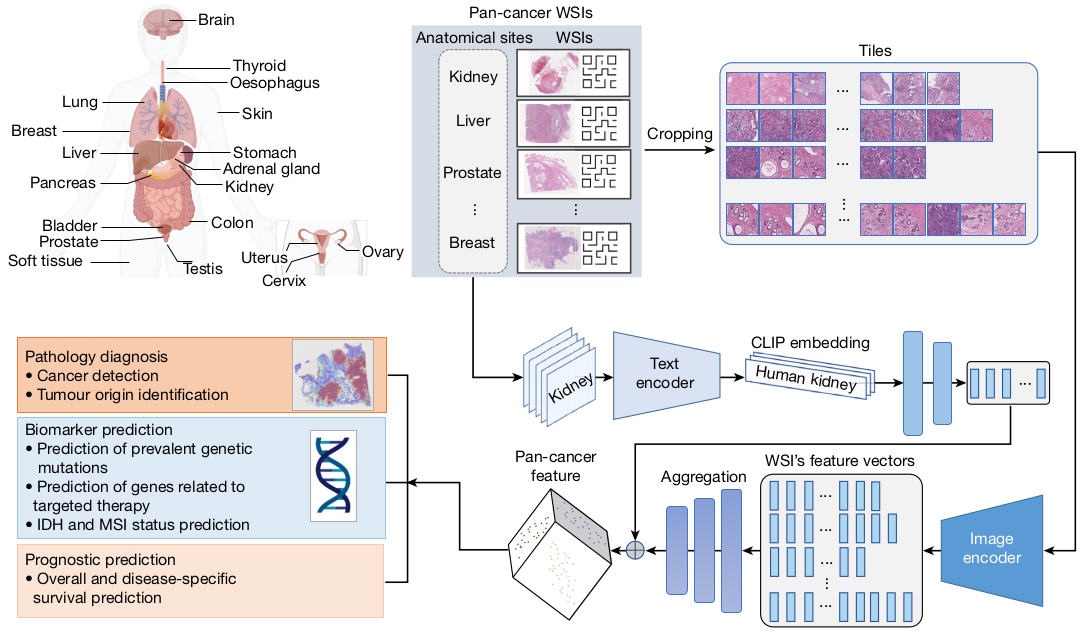
Developed as a general-purpose pathology AI system, CHIEF aims to accommodate a wide range of tissue types and evaluation tasks. Unlike conventional AI models that focus on task-specific learning, CHIEF employs a weakly supervised machine learning framework to extract pathology imaging features for systematic cancer evaluation. This allows CHIEF to generalize across different types of cancer and histopathological datasets, making it a versatile and scalable solution.
How CHIEF Works
CHIEF leverages two complementary methods of AI model pretraining to extract diverse pathology representations:
- Self-supervised pretraining: Using 15 million pathology image tiles, CHIEF learns tile-level feature representations without human annotations.
- Weakly supervised pretraining: Trained on 60,530 WSIs spanning 19 anatomical sites, CHIEF gains a comprehensive understanding of tissue context.
Additionally, CHIEF incorporates anatomical site information encoding to enhance the supervision process. By embedding anatomic site information using a contrastive language–image pretraining (CLIP) model, CHIEF improves its ability to differentiate between different tissue types.
Data Processing and Feature Encoding
One of the key strengths of CHIEF is its advanced data preprocessing and feature encoding techniques. Histopathological images are broken down into tiles using threshold-based methods like Otsu’s and Triangle’s thresholds. Each tile is then analyzed to extract relevant features, ensuring that the model captures meaningful tissue structures.
For feature encoding, CHIEF uses:
- CTransPath, a pre-trained transformer model, to quantify each tile’s representation.
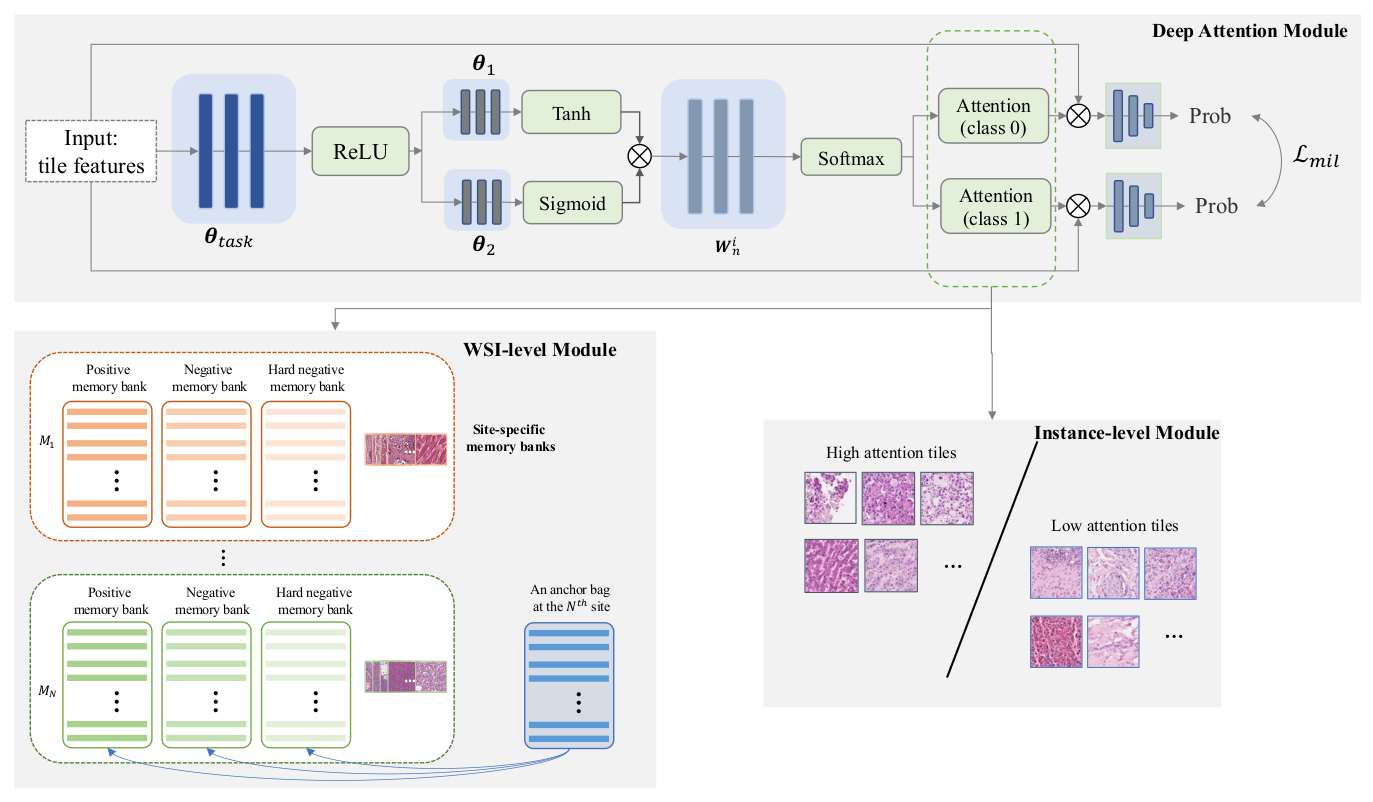
- A feature aggregation network, which integrates contextual information across tiles within each WSI.
- An attention-based pooling strategy, consisting of three modules designed to focus on the most relevant areas of the image.
Evaluation: Measuring CHIEF’s Performance
To validate its effectiveness, CHIEF was tested on four critical WSI-level prediction tasks:
- Cancer cell detection – Differentiating between malignant and benign samples.
- Tumor origin identification – Classifying tumors based on their anatomical site.
- Genomic profile characterization – Predicting genetic mutations associated with cancer.
- Survival outcome prediction – Estimating patient survival rates based on histopathology features.
For these tasks, CHIEF’s pretrained weights were fine-tuned with additional task-specific layers, demonstrating its flexibility in adapting to various histopathological challenges.

Results: Outperforming State-of-the-Art AI Models
CHIEF’s results were groundbreaking. It significantly outperformed existing deep learning models by up to 36%, particularly in handling domain shifts caused by differences in patient populations and slide preparation methods. Key performance metrics included:
- High AUROC scores in cancer classification and genomic profile prediction tasks.
- Accurate survival predictions, with strong concordance index (c-index) values.
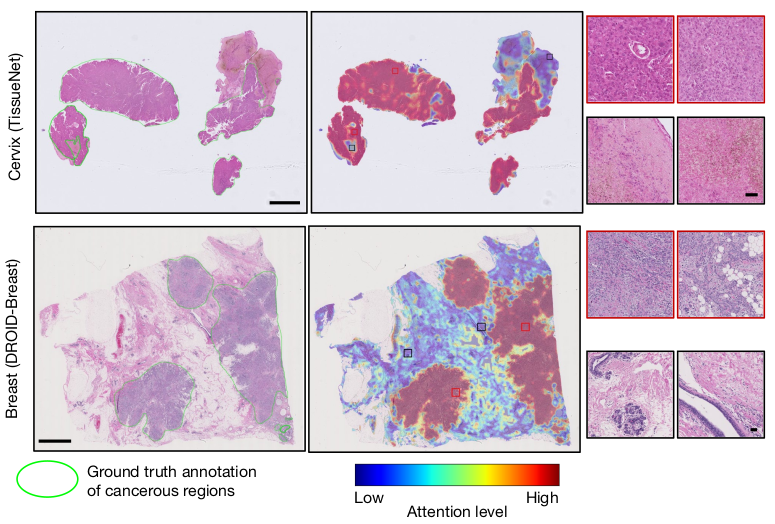
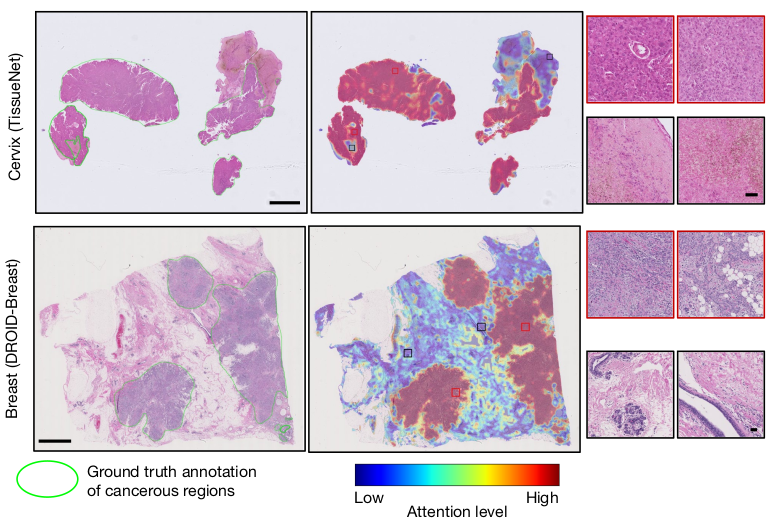
- Effective model attention visualization, showing CHIEF’s ability to pinpoint cancerous regions in WSIs accurately.
Future Prospects
The success of CHIEF highlights the potential for foundation models in pathology. Future research aims to:
- Incorporate more non-malignant slides and rare disease cases to further enhance the model’s generalizability.
- Extend its capabilities to evaluate the efficacy of novel cancer treatments, aiding in drug discovery and precision medicine.
Conclusion
The CHIEF foundation model represents a paradigm shift in AI-driven cancer diagnostics. By leveraging massive datasets, self-supervised learning, and advanced feature encoding techniques, CHIEF not only improves diagnostic accuracy but also enhances our understanding of cancer pathology. As research continues, foundation models like CHIEF are poised to transform cancer care, offering more reliable and scalable solutions for oncologists and pathologists worldwide.
The future of cancer diagnosis is here—and it’s powered by AI.
References
- WANG, Xiyue, ZHAO, Junhan, MAROSTICA, Eliana, et al. A pathology foundation model for cancer diagnosis and prognosis prediction. Nature, 2024, vol. 634, no 8035, p. 970-978.
- CHIEF Model GitHub Repository: https://github.com/hms-dbmi/CHIEF
