Uncertainty Quantification for skin cancer classification using Bayesian Deep Learning
Introduction
In this blogpost, we will present an interesting paper by Abdar et al. published in 2021 in Computers in Biology and Medecine, which is "Uncertainty quantification in skin cancer classification using three-way decision-based Bayesian deep learning".
The use of artificial intelligence (AI) in healthcare has grown exponentially, offering powerful tools to assist in disease diagnosis. Among these applications, skin cancer classification has gained significant attention, thanks to its potential to save lives through early and accurate detection. However, a critical question remains: can AI systems be trusted to make reliable decisions in high-stakes scenarios like medical diagnosis?
This is where uncertainty quantification (UQ) enters the conversation. In their paper, Abdar et al. propose a solution to control uncertainty in AI-powered skin cancer detection by combining Bayesian deep learning and a novel decision-making framework called the Three-Way Decision (3WD) approach. This combination not only improves accuracy but also introduces a layer of safety and accountability to AI systems in healthcare.
Why Uncertainty Quantification Matters
Medical AI systems often function in environments where the consequences of a wrong decision can be dire. For instance, a misclassification in skin cancer detection could lead to delayed treatment or unnecessary procedures. Unlike traditional AI models, which offer a single prediction, models with UQ provide insight into how confident the model is in its decision.
Two key types of uncertainty are addressed:
- Aleatoric Uncertainty: Uncertainty due to inherent noise in the data (e.g., low-quality images).
- Epistemic Uncertainty: Uncertainty stemming from the model itself, often due to limited training data or unseen cases.
By quantifying these uncertainties, the AI system can make informed decisions about whether it should act on its prediction or defer the decision to a medical expert.
The Power of Bayesian Deep Learning
Monte Carlo Dropout
In [1], the authors proposed a bayesian theory of the dropout. They showed that optimizing a NN by dropout (as a regularization method) can be equivalent to a type of variational inference of a Bayesian ML model.
In traditional dropout, neurons are randomly "dropped" (set to zero) during training to prevent overfitting. However, dropout is typically disabled during inference., In MC Dropout, dropout remains active during inference, effectively treating the model as a stochastic ensemble and approximating the posterior distribution over the model's weights.
Deep Ensemble
[2] proposed to use Deep Ensemble Model as a simple and scalable alternative to Bayesian NNs. Creating a Bayesian model with Deep Ensembles involves training multiple neural networks independently and using their combined predictions to approximate uncertainty. This approach is straightforward, powerful, and avoids the complexities of fully Bayesian neural networks.
Ensemble Monte Carlo Dropout
In [3] the authors suggest to create a Bayesian model with Ensemble Monte Carlo (MC) Dropout that combines the strengths of two uncertainty quantification methods: Deep Ensembles and MC Dropout. This approach uses multiple models (ensemble) with dropout layers enabled during both training and inference.
These three proposed methods are simple and scalable, that is why the authors mainly focused on their use, but it is important to notice that it exists many other methods to help measure uncertainty. It is also worth saying that Bayesian methods are computationally much more expensive than traditional Deep learning methods.
The Three-Way Decision Framework
The authors use 3WD in conjunction with Bayesian deep learning to manage uncertain predictions in skin cancer classification. Here's how it works:
1. Measuring Uncertainty
- Uncertainty is quantified using entropy H of the predictive distribution.
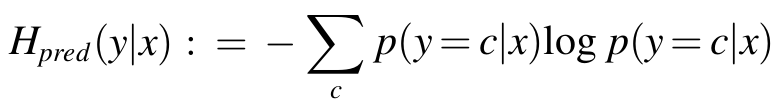
- High entropy indicates low confidence in the prediction, and vice versa.
2. Applying Three Decision Options
Based on the entropy, the model makes one of these decisions:
- Accept: If entropy is low, the model is confident in its prediction and directly classifies the input as benign or malignant.
- Defer: If entropy falls within an intermediate range, the model defers the decision for further investigation. This could involve:
- Acquiring additional data (e.g., more images or patient history).
- Escalating the case to a medical expert.
3. Threshold Setting
- The threshold for accept or defer is determined during model training or validation by analyzing the entropy distribution of predictions.
- This threshold is optimized to minimize overall risk and ensure the safety of decisions.

Conclusion: A Safer Path Forward for AI in Healthcare
The work by Abdar et al. highlights the importance of moving beyond traditional accuracy metrics in AI models. By embracing uncertainty quantification and implementing safeguards like the Three-Way Decision framework, we can create AI systems that are more aligned with the complex realities of medical diagnostics.
This research paves the way for AI systems that work collaboratively with humans, augmenting their capabilities while deferring to their expertise when needed. It’s an exciting step forward for AI in healthcare, and a reminder that responsible AI development is just as important as technical innovation.
References
[1] Y. Gal, Z. Ghahramani, Dropout as a bayesian approximation: representing model uncertainty in deep learning, in International Conference on Machine Learning, 2016.
[2] B. Lakshminarayanan, A. Pritzel, C. Blundell, Simple and scalable predictive uncertainty estimation using deep ensembles, in Advances in Neural Information Processing Systems, 2017
[3] A. Filos,et al. A systematic comparison of bayesian deep learning robustness in diabetic retinopathy tasks, in NIPS Bayesian Deep Learning Workshop, 2019
