Computational Protein Design with Automated Reasoning and Deep Learning
Marianne DEFRESNE
PhD, AI Scientist at Torus AI
February 8th, 2024
Abstract
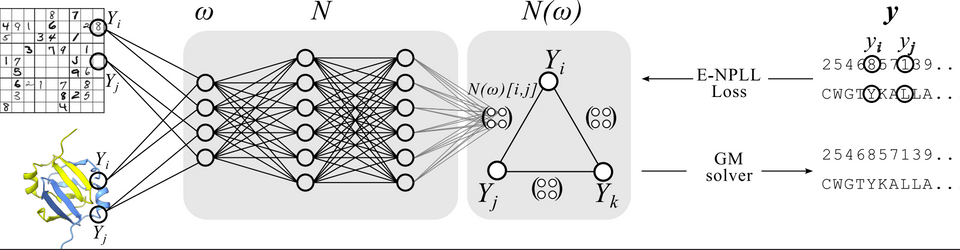
Proteins are complex molecules that perform many functions in living organisms. Some of these functions can be repurposed for applications in biotechnology, medicine, and green chemistry... The goal of Computational Protein Design (CPD) is to predict a protein sequence fit for an application. Since the function of a protein is tightly linked to its 3D structure, CPD can be formulated as predicting a sequence folding onto a target structure and therefore fulfilling a function of interest. Existing approaches are based on the optimization of an energy function scoring interactions within the proteins or they are purely based on Deep Learning. In this thesis, we present a new hybrid approach for CPD, combining Deep Learning (DL) and Automated Reasoning. Our first contribution is to categorize existing DL approaches based on protein representation. The discussion of their advantages and drawbacks with respect to traditional energy-based methods leads us to try and take the best of both worlds by learning a new scoring function that is optimized to design proteins. This score function is a Graphical Model, a reasoning compound that has already successfully been used to optimize proteins. This objective requires a hybrid pipeline combining Deep Learning and discrete optimization. Such hybridization being an open challenge in Artificial Intelligence, we first developed a method to learn Graphical Models from data that allows exact inference while being scalable. It was developed on the standard benchmark of learning how to play Sudoku, in which it achieves state-of-the-art results. We then applied this hybrid pipeline to protein design. A protein structure being non-Euclidean data, it requires a suited representation and a fitting neural architecture to be processed. We learned a new scoring function for design that we named Effie. We extensively validated it in silico. On design tasks, it outperformed traditional energy-based methods while being competitive with DL-based approaches. Moreover, it can tackle tasks for which it has not been explicitly trained, suggesting that some physical-chemical concepts have been learned. Finally, we applied it on 3 projects where the design objectives required to bias or conditioned Effie a posteriori via the addition of knowledge or constraints. In this context, we showed the interest of our hybrid approach as Effie + discrete optimization outperformed pure Deep Learning methods.
Supplementary Materials
Papers
- https://www.mdpi.com/1422-0067/22/21/11741
- https://www.ijcai.org/proceedings/2023/0402.pdf
Web Pages
- https://aihub.org/2023/06/07/bridging-the-gap-between-learning-and-reasoning/
